Research
Our group uses the fruit fly Drosophila melanogaster—whose brain is compact (~0.5 mm) and amenable to powerful genetic manipulation. We are especially interested in how regulations at the level of protein translation shape neural function and behavior.
Why Drosophila?
Drosophila is an exceptional model to ask "how the brains function and change?". Brain-wide wiring has been mapped at electron-microscopy resolution, detailing the network architecture. These flies exhibit rich repertoire of behaviors that can be modulated by past-experiences, including associative learning (linking an odor to sugar reward or electric shock), escalation of alcohol consumption with repeated exposure, or suppression of courtship after rejection. With the help of highly-sophisticated genetic toolbox, we are able to investigate functions of nearly every gene and cell type.
Why study translation?
Tuned expression of genetic code is the basis of neural functions and plasticity. While recent transcriptome analyses have revealed extensive transcriptional control, accumulating evidence suggests that regulation at the level of translation is also crucial. Disruption of translational regulators, such as FMR1, the mTOR pathway, and eIF2αlpha kinases, has been associated with neurodevelopmental and neuropsychiatric conditions. Actually, studies comparing transcriptome and proteome suggest that mRNA abundance explains only ~30–40% of protein levels. Translational regulations can even modulate sequences of polypeptides, through multiple translation start sites or readthrough of stop codons. Therefore, translational regulation is one of the key frontiers in the field of neurobiology.
We integrate high-precision behavioral measurements and neuroanatomy with genome-wide measurement of protein synthesis.
Behavior, genetics, and anatomy
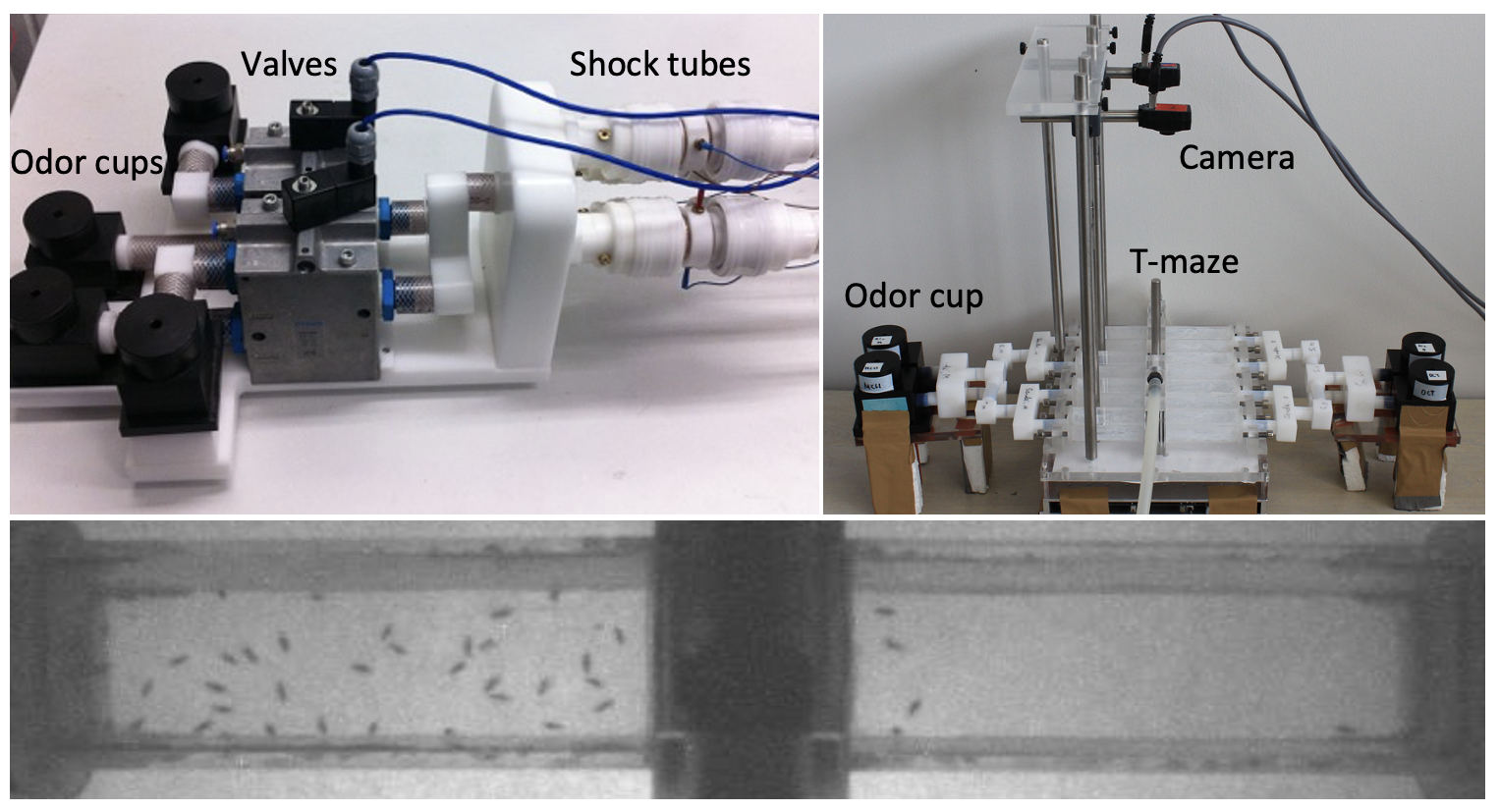
We built custom-made, high-throughput rigs to measure olfactory learning and feeding choices involving alcohol, psychostimulants, or toxins (Fig. 1–2). Through transgenesis and genome editing, we can visualize genetically defined neurons and specific molecules in the brain (Fig. 3).
Genome-wide translation profiling
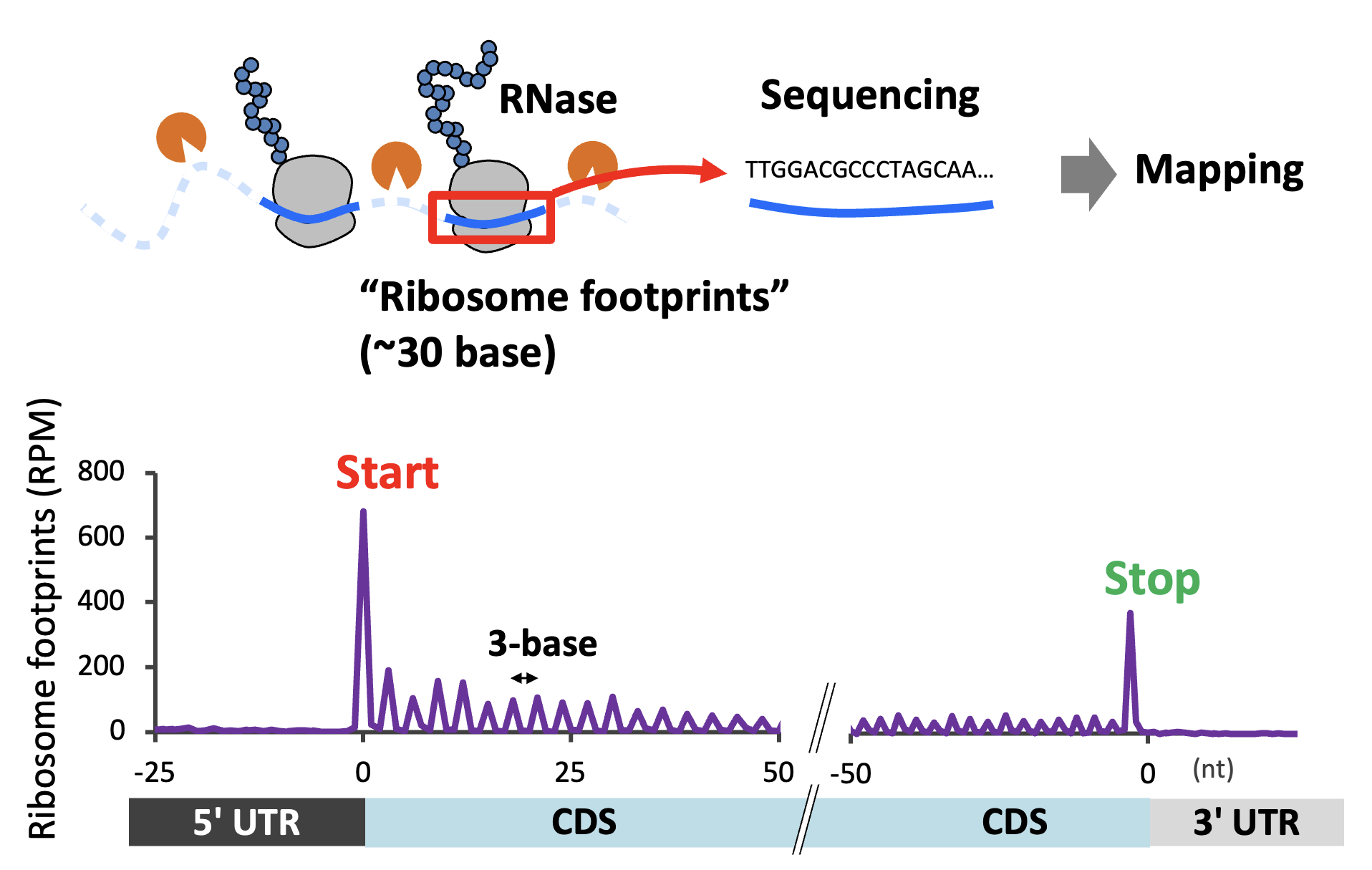
Ribosome profiling (Ribo-seq) maps ribosomes on mRNAs genome-wide to read out translation directly. By pairing transgenic tagging in flies with Ribo-seq, we are analyzing translation from targeted brain cell types (Fig. 4).
Using these approaches, we are dissecting how physiological stimuli, such as associative learning, drug/food intake, or neuronal activity, reprogram translation and how this reprogramming impacts behaviors. We are also interested in the functions and molecular mechanisms of unconventional translation in the nervous system.